Code
source("scripts/99-pkgs.R")Explore the correlation between plant cover estimated by RPAS (RPAS-estimated vegetation cover) and estimated by field measures (Field-estimated vegetation cover)
Compute the RMSE (Root Mean Squared Error) and the normalized RMSE
| RMSE | min | max | norm. RMSE (%) |
|---|---|---|---|
| 9.89 | 6 | 87 | 12.21 |
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | -2.868642 | 1.8882404 | -1.519214 | 0.132065 |
| plant_coverage_field | 1.124453 | 0.0537794 | 20.908615 | 0.000000 |
| r.squared | 0.8230322 |
| adj.r.squared | 0.8211496 |
| sigma | 9.6840794 |
| statistic | 437.1701925 |
| p.value | 0.0000000 |
| df | 1.0000000 |
| logLik | -353.1739261 |
| AIC | 712.3478521 |
| BIC | 720.0408967 |
| deviance | 8815.4509553 |
| df.residual | 94.0000000 |
| nobs | 96.0000000 |
The p-value of the regression is 4.0417611^{-37}.
# https://fhernanb.github.io/libro_regresion/diag2.html#estad%C3%ADstica-press-y-r2-de-predicci%C3%B3n
# https://rpubs.com/RatherBit/102428
# https://tomhopper.me/2014/05/16/can-we-do-better-than-r-squared/
PRESS <- function(linear.model) {
# calculate the predictive residuals
pr <- residuals(linear.model) / (1 - lm.influence(linear.model)$hat)
# calculate the PRESS
PRESS <- sum(pr^2)
return(PRESS)
}
pred_r_squared <- function(linear.model) {
#' Use anova() to get the sum of squares for the linear model
lm.anova <- anova(linear.model)
#' Calculate the total sum of squares
tss <- sum(lm.anova$"Sum Sq")
# Calculate the predictive R^2
pred.r.squared <- 1 - PRESS(linear.model) / (tss)
return(pred.r.squared)
}# https://stackoverflow.com/questions/17022553/adding-r2-on-graph-with-facets
lm_r2 <- function(df) {
m <- lm(plant_coverage_rpas ~ plant_coverage_field, df)
eq <- substitute(
r2,
list(r2 = format(summary(m)$r.squared, digits = 3))
)
as.character(as.expression(eq))
}
# lm_eqn = function(df, model){
# eq <- substitute(
# italic(y) == a + b %.% italic(x) * "," ~ ~pvalue~p,
# list(
# a = format(coef(model)[1], digits = 3),
# b = format(coef(model)[2], digits = 3),
# p = ifelse(
# summary(m)$coefficients[2,'Pr(>|t|)'] < 0.0001,
# "< 0.0001",
# "= ~format(summary(m)$coefficients[2,'Pr(>|t|)'])"
# )
# ))
# as.character(as.expression(eq))
#
# }verde_claro <- "#c6ddb3"
verde_oscuro <- "#3e6c62"
ylab <- "Drone-estimated"
xlab <- "Field-estimated"
# See https://stackoverflow.com/questions/65076492/ggplot-size-of-annotate-vs-size-of-element-text
# Custom theme
theme_rpas <- function() {
theme_bw() %+replace%
theme(
plot.title = element_text(
size = 14,
margin = margin(0, 0, 10, 0)
),
panel.grid = element_blank(),
axis.title = element_text(size = 12),
# For panels
strip.background = element_rect(fill = "white"),
strip.text = element_text(
size = 10,
margin = margin(5, 0, 5, 0)
)
)
}formula <- y ~ x
general_plot <- df %>%
ggplot(aes(x = plant_coverage_field, y = plant_coverage_rpas)) +
geom_point(size = 3, alpha = .5, colour = verde_oscuro) +
geom_abline(slope = 1, colour = verde_oscuro) +
labs(
x = xlab, y = ylab,
title = "Vegetation cover (%)"
) +
xlim(0, 100) +
ylim(0, 100) +
stat_poly_eq(
formula = formula,
aes(label = paste(
after_stat(eq.label), "*\"; \"*",
after_stat(adj.rr.label), "*\"; \"*",
after_stat(p.value.label), "*\"\"",
sep = ""
)),
rr.digits = 3,
colour = "black",
size = 8 / .pt
) +
annotate("text",
x = 0, y = 85,
label = paste0("RMSE = ", round(df.rmse_global$rmse, 2)),
colour = "black",
size = 12 / .pt,
hjust = 0
) +
annotate("text",
x = 0, y = 75,
label = paste0(
"italic(R)[predic.]^2~'='~",
round(pred_r_squared(m), 3)
),
colour = "black",
parse = TRUE,
size = 12 / .pt,
hjust = 0
) +
theme_rpas() +
geom_smooth(method = "lm", col = "gray", se = FALSE, size = 1)
general_plot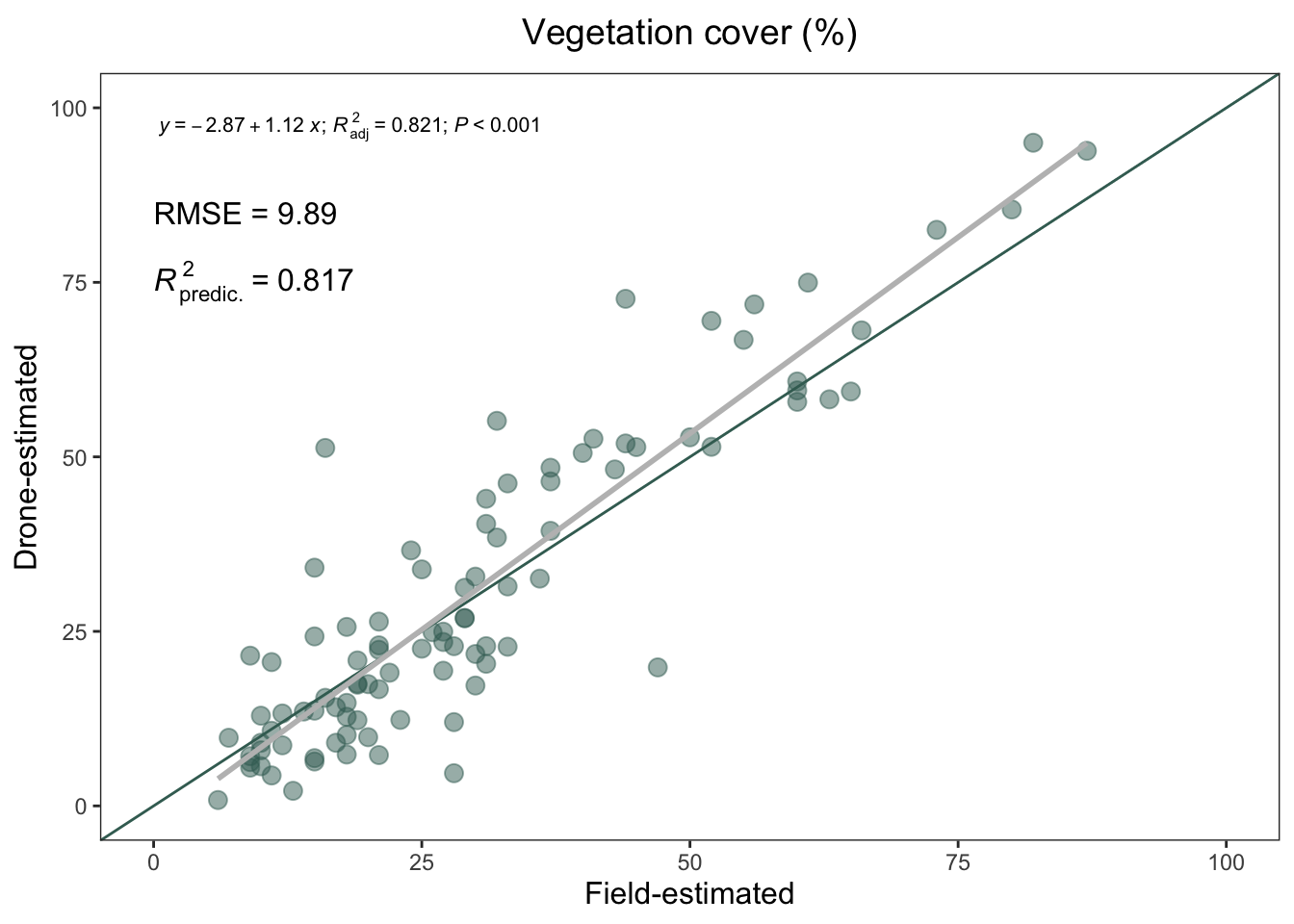
df.rmse_treatment <- df %>%
group_by(treatment) %>%
summarise(
rmse = round(
Metrics::rmse(plant_coverage_field, plant_coverage_rpas), 4
),
min = min(plant_coverage_field),
max = max(plant_coverage_field),
rmsen.minmax = rmse / (max(plant_coverage_field) - min(plant_coverage_field)) * 100
)
# Compute predictive R2
df.rmse_treatment <-
df.rmse_treatment |> inner_join(
df |> group_by(treatment) |>
group_modify(~ data.frame(
pred.r2 = pred_r_squared(
lm(plant_coverage_rpas ~ plant_coverage_field, data = .x))))
)| Treatment | RMSE | min | max | norm. RMSE (%) | pred R2 |
|---|---|---|---|---|---|
| Prescribed Burn | 7.95 | 9 | 66 | 13.95 | 0.748 |
| Pyric Herbivorism | 10.73 | 6 | 87 | 13.24 | 0.832 |
treatment_plot <- df %>%
ggplot(aes(x = plant_coverage_field, y = plant_coverage_rpas)) +
geom_point(size = 3, alpha = .5, colour = verde_oscuro) +
geom_abline(slope = 1, colour = verde_oscuro) +
facet_wrap(~treatment, labeller = label_value) +
labs(x = xlab, y = ylab, title = "Vegetation cover (%)") +
xlim(0, 100) +
ylim(0, 100) +
theme_rpas() +
theme(legend.position = "none") +
geom_richtext(
data = df.rmse_treatment, size = 8 / .pt,
aes(
x = 0, y = 80,
label = paste0(
"RMSE<sub>norm.</sub> = ",
round(rmsen.minmax, 2), " %"
),
hjust = 0
),
fill = NA, label.color = NA
) +
stat_poly_eq(
formula = formula,
aes(label = paste(
after_stat(adj.rr.label), "*\"; \"*",
after_stat(p.value.label), "*\"\"",
sep = ""
)),
rr.digits = 3,
colour = "black",
size = 8 / .pt
) +
geom_richtext(
data = df.rmse_treatment, size = 8 / .pt,
aes(
x = 0, y = 70,
label = paste0(
"*R*<sup>2</sup><sub>predic.</sub> = ",
round(pred.r2, 3)
),
hjust = 0
),
fill = NA, label.color = NA
) +
geom_smooth(method = "lm", col = "gray", se = FALSE, size = 1)
treatment_plot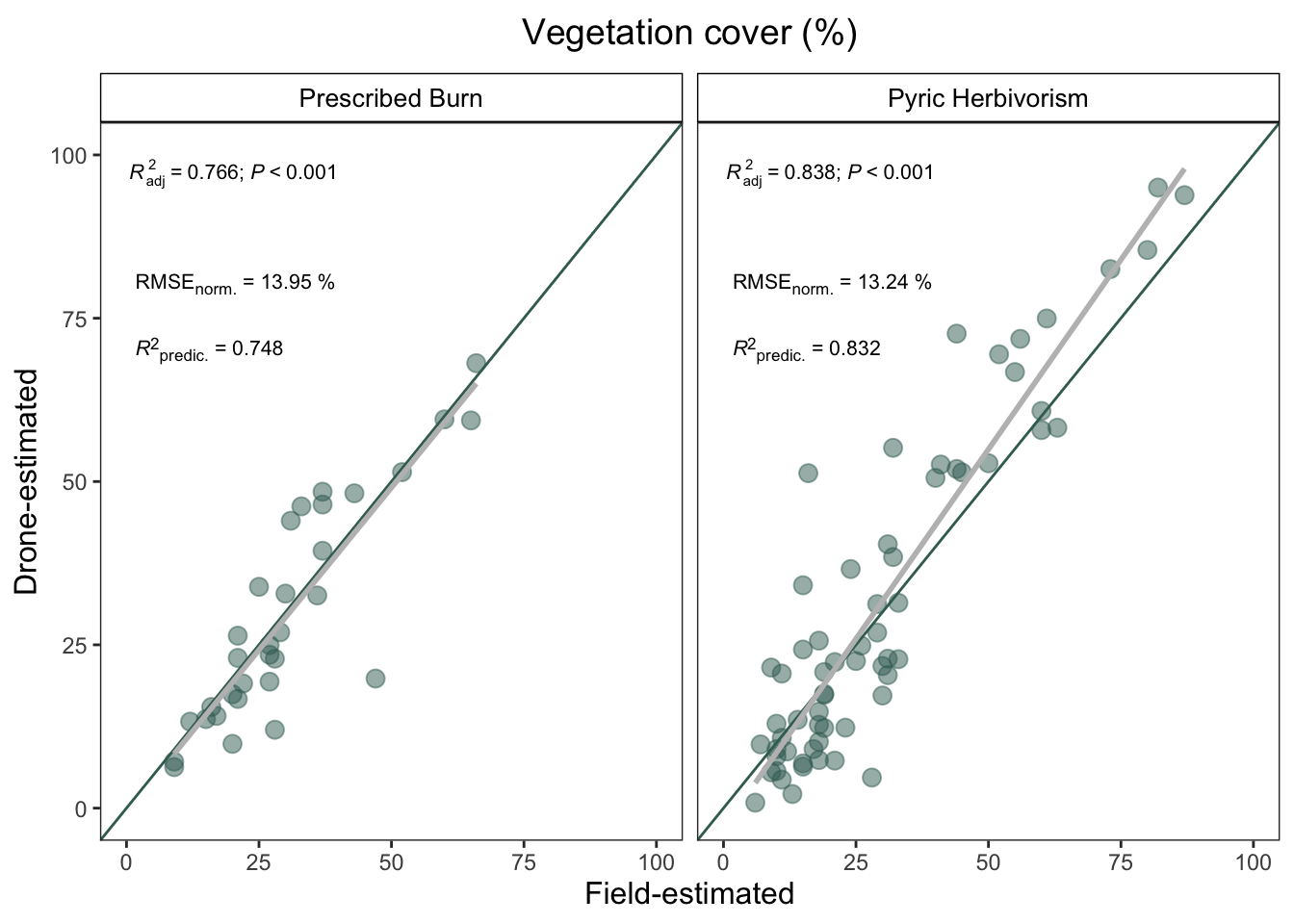
df.rmse_groups <- df %>%
group_by(cover_type) %>%
summarise(
rmse = round(
Metrics::rmse(plant_coverage_field, plant_coverage_rpas), 4
),
min = min(plant_coverage_field),
max = max(plant_coverage_field),
rmsen.minmax = rmse / (max(plant_coverage_field) - min(plant_coverage_field)) * 100
)
# Compute predictive R2
df.rmse_groups <-
df.rmse_groups |> inner_join(
df |> group_by(cover_type) |>
group_modify(~ data.frame(
pred.r2 = abs(pred_r_squared(
lm(plant_coverage_rpas ~ plant_coverage_field, data = .x)))))
)| Cover type | RMSE | min | max | norm. RMSE (%) | pred R2 |
|---|---|---|---|---|---|
| High-cover alfa grass steppe | 7.63 | 9 | 87 | 9.78 | 0.938 |
| High-cover shrubland | 12.69 | 10 | 61 | 24.89 | 0.571 |
| Low-cover shrubland | 7.30 | 6 | 28 | 33.18 | 0.145 |
| Moderate-cover shrubland | 10.89 | 9 | 63 | 20.16 | 0.469 |
custom_order <- c(
"Low-cover shrubland", "Moderate-cover shrubland",
"High-cover shrubland", "High-cover alfa grass steppe"
)
covertype_plot <- df %>%
ggplot(aes(x = plant_coverage_field, y = plant_coverage_rpas)) +
geom_point(size = 3, alpha = .5, colour = verde_oscuro) +
geom_abline(slope = 1, colour = verde_oscuro) +
facet_wrap(~ factor(cover_type, custom_order),
labeller = label_value
) +
labs(x = xlab, y = ylab, title = "Vegetation cover (%)") +
xlim(0, 100) +
ylim(0, 100) +
theme_rpas() +
theme(legend.position = "none") +
geom_richtext(
data = df.rmse_groups, size = 8 / .pt,
aes(
x = 0, y = 80,
label = paste0(
"RMSE<sub>norm.</sub> = ",
round(rmsen.minmax, 2), " %"
)
),
hjust = 0,
fill = NA, label.color = NA
) +
stat_poly_eq(
formula = formula,
aes(label = paste(
after_stat(adj.rr.label), "*\"; \"*",
after_stat(p.value.label), "*\"\"",
sep = ""
)),
rr.digits = 3,
colour = "black",
size = 8 / .pt
) +
geom_richtext(
data = df.rmse_groups, size = 8 / .pt,
aes(
x = 0, y = 70,
label = paste0(
"*R*<sup>2</sup><sub>predic.</sub> = ",
round(pred.r2, 3)
),
hjust = 0
),
fill = NA, label.color = NA
) +
geom_smooth(method = "lm", col = "gray", se = FALSE, size = 1)
covertype_plot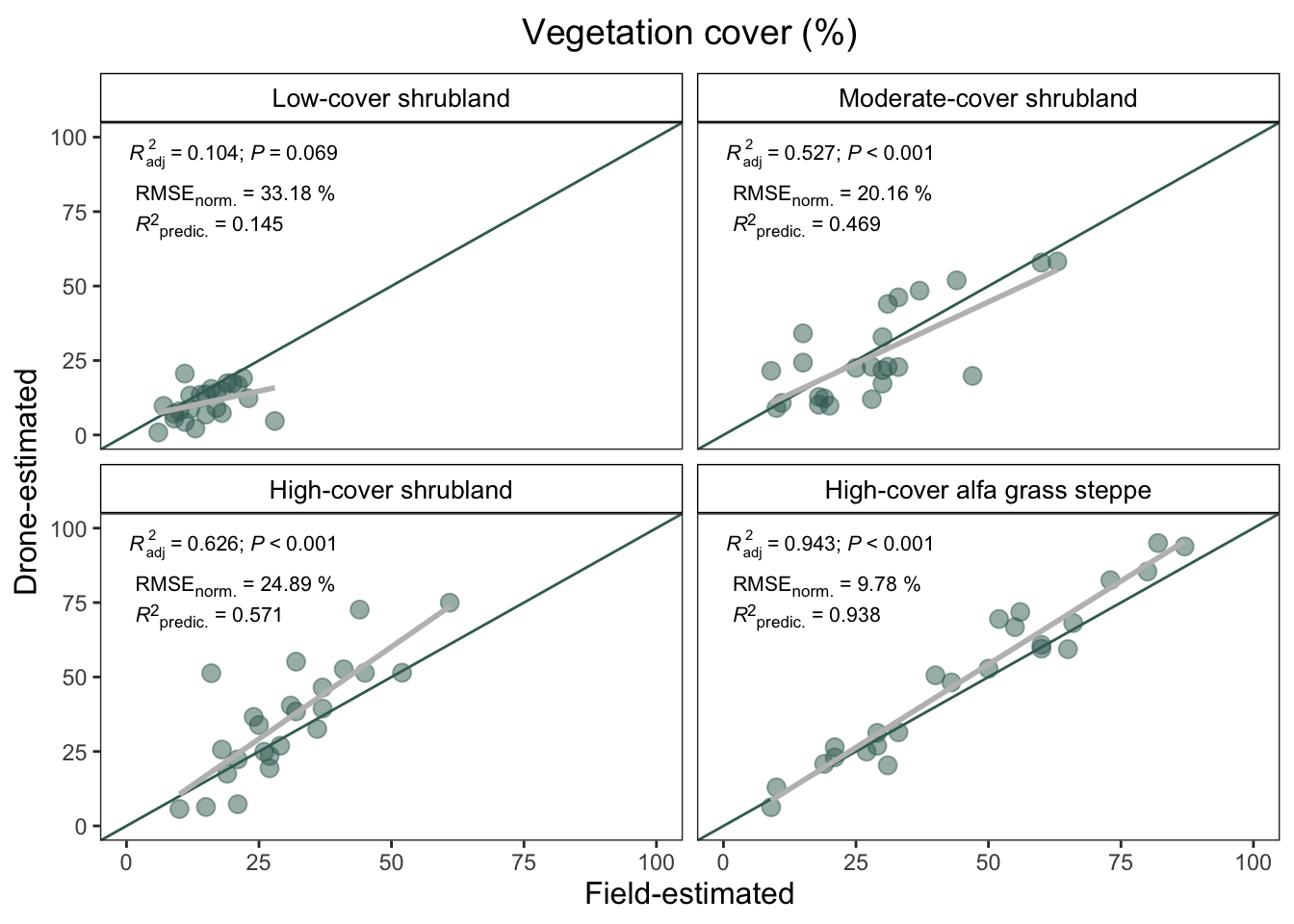
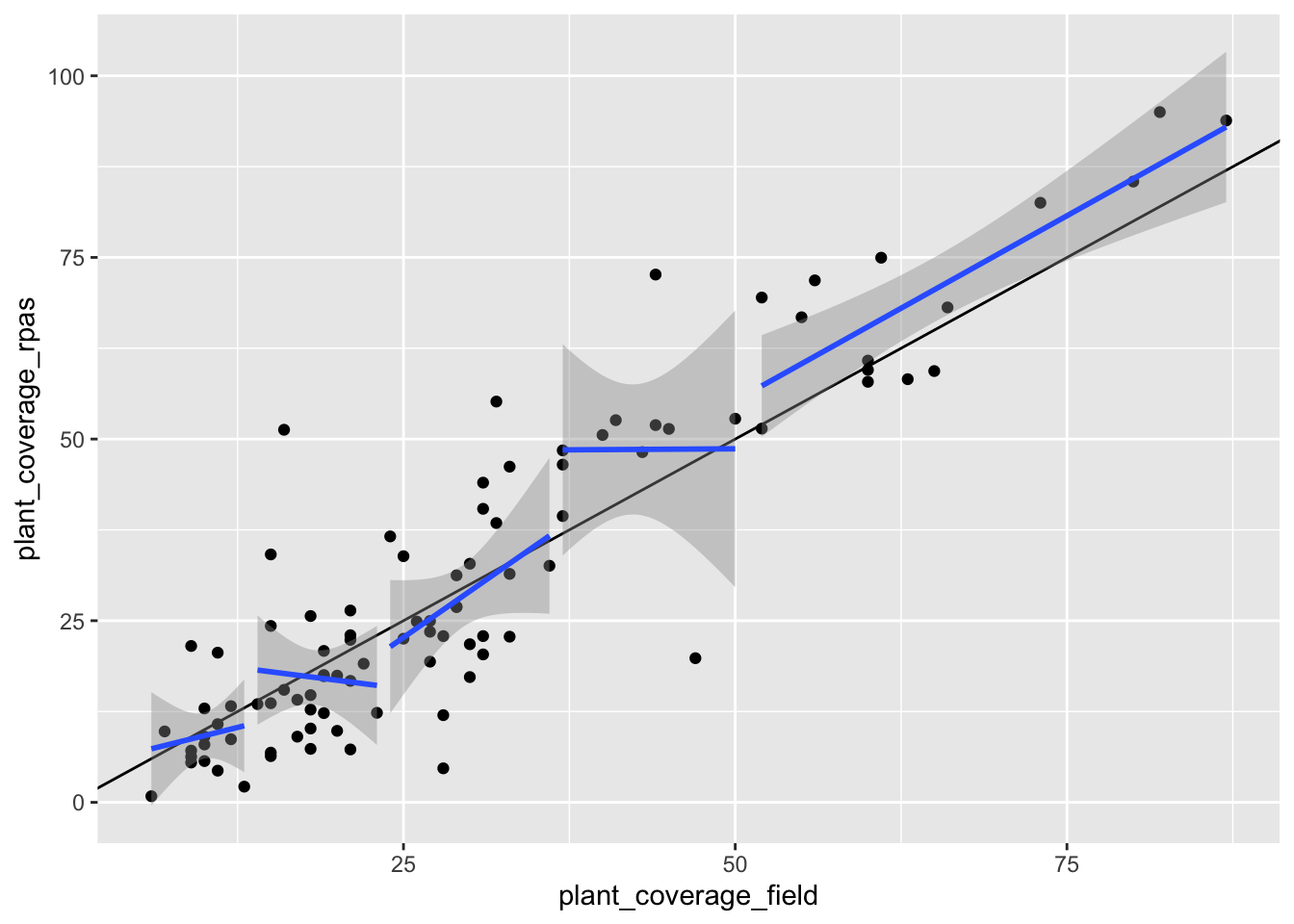
Is the relationship between the vegetation coverage estimated by RPAS and by field measurement uniform across all coverage values? or is the correlation between those two approach homogeneous across all coverage values?
Model formula:
plant_coverage_rpas ~ plant_coverage_field
Fitted party:
[1] root
| [2] plant_coverage_field <= 36
| | [3] plant_coverage_field <= 23
| | | [4] plant_coverage_field <= 13: 9.150 (n = 16, err = 498.4)
| | | [5] plant_coverage_field > 13: 17.207 (n = 28, err = 2433.9)
| | [6] plant_coverage_field > 23: 28.322 (n = 26, err = 2998.9)
| [7] plant_coverage_field > 36
| | [8] plant_coverage_field <= 50: 48.571 (n = 11, err = 1550.7)
| | [9] plant_coverage_field > 50: 70.347 (n = 15, err = 2566.3)
Number of inner nodes: 4
Number of terminal nodes: 5$`1`
plant_coverage_field
statistic 7.818806e+01
p.value 9.368296e-19
$`2`
plant_coverage_field
statistic 2.791016e+01
p.value 1.270809e-07
$`3`
plant_coverage_field
statistic 5.75223821
p.value 0.01646766
$`4`
NULL
$`5`
plant_coverage_field
statistic 0.1017646
p.value 0.7497222
$`6`
plant_coverage_field
statistic 2.83932243
p.value 0.09198299
$`7`
plant_coverage_field
statistic 1.604824e+01
p.value 6.174895e-05
$`8`
NULL
$`9`
NULLThere are 5 terminal nodes.
ggparty(ct) +
geom_edge() +
geom_edge_label(colour = "grey", size = 4) +
geom_node_plot(
gglist = list(
geom_point(aes(x = plant_coverage_field, y = plant_coverage_rpas)),
geom_smooth(aes(x = plant_coverage_field, y = plant_coverage_rpas), method = lm, se = FALSE),
geom_abline(slope = 1, colour = "gray"),
theme_bw(base_size = 10),
xlab(xlab),
ylab(ylab)
),
scales = "fixed",
shared_axis_labels = TRUE,
shared_legend = TRUE,
legend_separator = TRUE,
id = "terminal"
) +
geom_node_label(aes(col = splitvar),
line_list = list(
aes(label = paste("Node", id)),
aes(label = splitvar),
aes(label = scales::pvalue(p.value,
accuracy = 0.001,
decimal.mark = ".",
add_p = TRUE
))
),
line_gpar = list(
list(size = 8, col = "black", fontface = "bold"),
list(size = 8),
list(size = 8)
),
ids = "inner"
) +
geom_node_label(aes(label = paste0("Node ", id, ", (n= ", nodesize, ")")),
fontface = "bold",
ids = "terminal",
size = 2,
nudge_y = 0.01
) +
theme(legend.position = "none")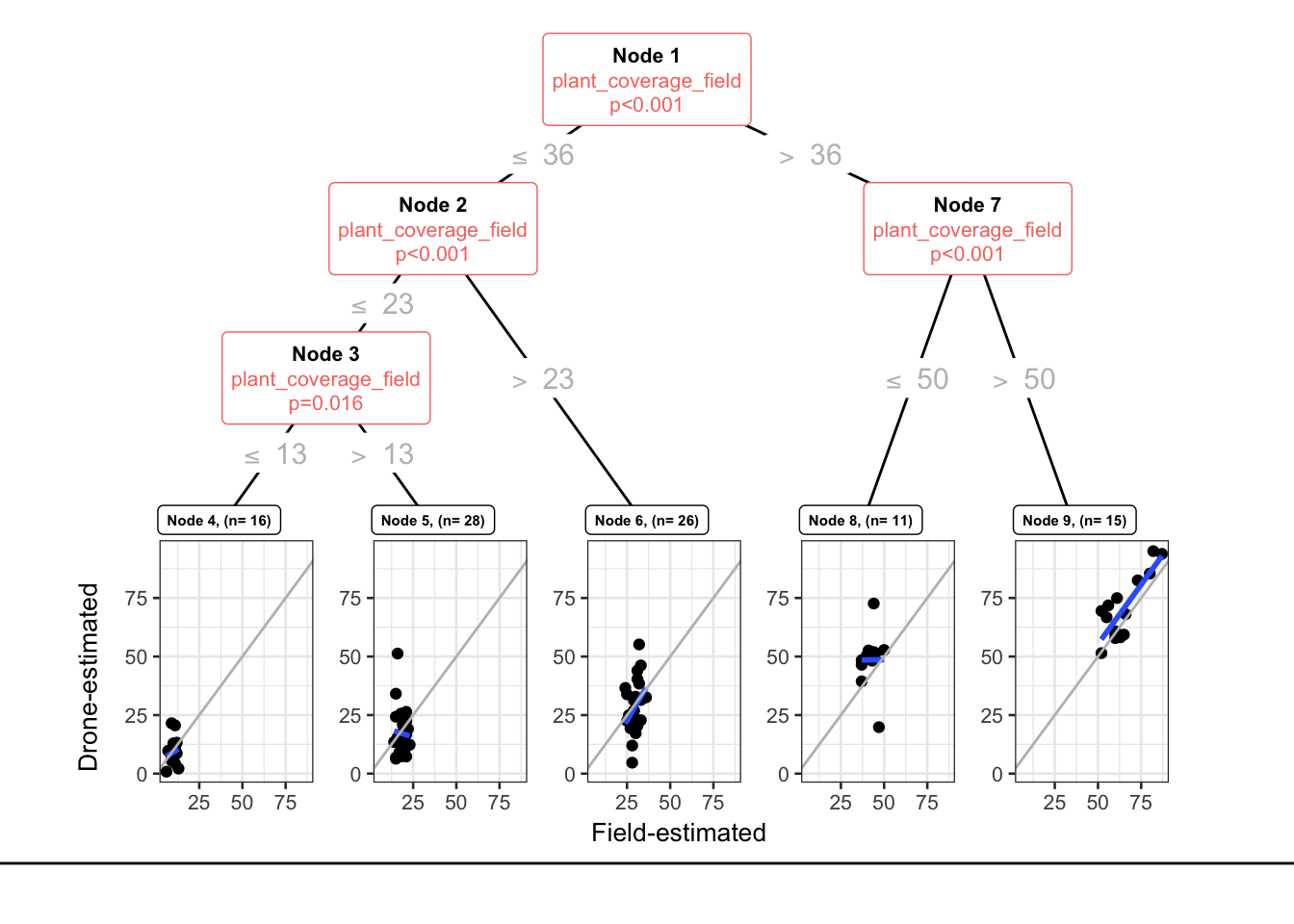
What about the overfitting?
ggplot(df, aes(x = plant_coverage_field, y = plant_coverage_rpas)) +
geom_point() +
geom_abline(yintercept = 1) +
geom_smooth(
data = (df %>% filter(plant_coverage_field <= 13)),
aes(x = plant_coverage_field, y = plant_coverage_rpas),
method = "lm"
) +
geom_smooth(
data = (df %>% filter(plant_coverage_field > 13 & plant_coverage_field <= 23)),
aes(x = plant_coverage_field, y = plant_coverage_rpas),
method = "lm"
) +
geom_smooth(
data = (df %>% filter(plant_coverage_field > 23 & plant_coverage_field <= 36)),
aes(x = plant_coverage_field, y = plant_coverage_rpas),
method = "lm"
) +
geom_smooth(
data = (df %>% filter(plant_coverage_field > 36 & plant_coverage_field <= 50)),
aes(x = plant_coverage_field, y = plant_coverage_rpas),
method = "lm"
) +
geom_smooth(
data = (df %>% filter(plant_coverage_field > 50)),
aes(x = plant_coverage_field, y = plant_coverage_rpas),
method = "lm"
)
n= 96
node), split, n, deviance, yval
* denotes terminal node
1) root 96 49813.8800 30.77125
2) plant_coverage_field< 36.5 70 9815.7370 19.49371
4) plant_coverage_field< 23.5 44 3593.3100 14.27727
8) plant_coverage_field< 13.5 16 498.4016 9.15000 *
9) plant_coverage_field>=13.5 28 2433.9300 17.20714 *
5) plant_coverage_field>=23.5 26 2998.9390 28.32154
10) plant_coverage_field< 30.5 16 998.4559 23.88250 *
11) plant_coverage_field>=30.5 10 1180.7530 35.42400 *
3) plant_coverage_field>=36.5 26 7126.2780 61.13385
6) plant_coverage_field< 51 11 1550.7140 48.57091 *
7) plant_coverage_field>=51 15 2566.3220 70.34667 *
Regression tree:
rpart(formula = plant_coverage_rpas ~ plant_coverage_field, data = df)
Variables actually used in tree construction:
[1] plant_coverage_field
Root node error: 49814/96 = 518.89
n= 96
CP nsplit rel error xerror xstd
1 0.659894 0 1.00000 1.02767 0.147971
2 0.064711 1 0.34011 0.39621 0.065669
3 0.060410 2 0.27540 0.38907 0.061846
4 0.016456 3 0.21499 0.29072 0.047114
5 0.013269 4 0.19853 0.28587 0.046682
6 0.010000 5 0.18526 0.26769 0.046179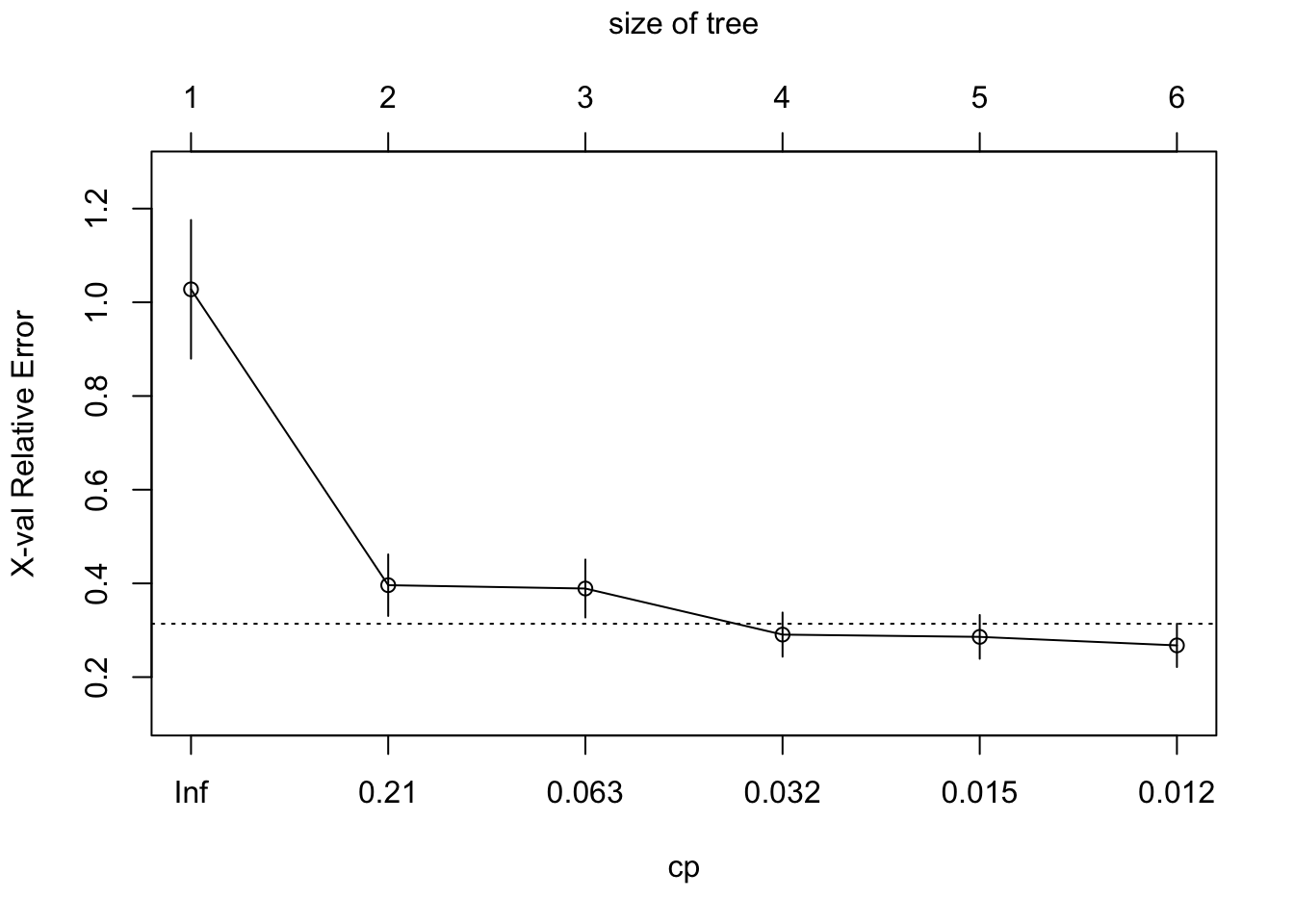
n= 96
node), split, n, deviance, yval
* denotes terminal node
1) root 96 49813.880 30.77125
2) plant_coverage_field< 36.5 70 9815.737 19.49371 *
3) plant_coverage_field>=36.5 26 7126.278 61.13385 *
Regression tree:
rpart(formula = plant_coverage_rpas ~ plant_coverage_field, data = df,
control = rpart.control(minsplit = 2, cp = 0.075))
Variables actually used in tree construction:
[1] plant_coverage_field
Root node error: 49814/96 = 518.89
n= 96
CP nsplit rel error xerror xstd
1 0.65989 0 1.00000 1.02767 0.147971
2 0.07500 1 0.34011 0.39621 0.065669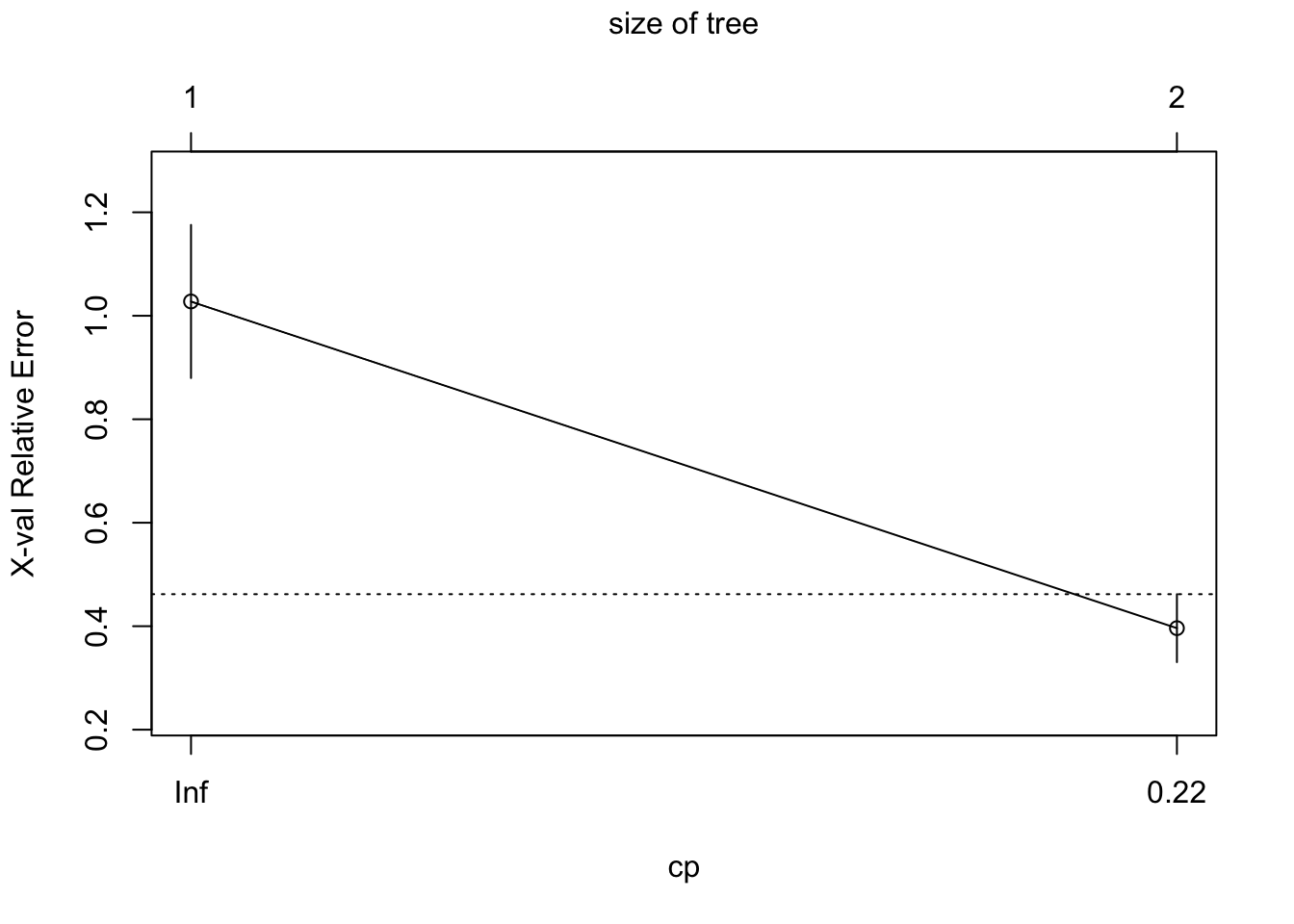
set.seed(123)
ct_ok <- partykit::ctree(plant_coverage_rpas ~ plant_coverage_field,
data = df,
control =
ctree_control(minsplit = 1, alpha = 0.05, maxdepth = 1)
)
plot_party <- ggparty(ct_ok) +
geom_edge() +
geom_edge_label(colour = verde_oscuro, size = 5) +
geom_node_plot(
gglist = list(
geom_point(
aes(
x = plant_coverage_field,
y = plant_coverage_rpas
),
size = 3, alpha = .5, colour = verde_oscuro
),
geom_smooth(aes(x = plant_coverage_field, y = plant_coverage_rpas),
method = lm, se = FALSE, colour = "gray"
),
geom_abline(slope = 1, colour = verde_oscuro),
theme_bw(base_size = 10),
xlab(xlab),
ylab(ylab),
theme(
panel.grid = element_blank()
)
),
scales = "fixed",
shared_axis_labels = TRUE,
shared_legend = TRUE,
legend_separator = TRUE,
id = "terminal"
) +
geom_node_label(aes(col = splitvar),
line_list = list(
aes(id),
aes(label = "Vegetation cover"),
aes(label = scales::pvalue(p.value,
accuracy = 0.001,
decimal.mark = ".",
add_p = TRUE
))
),
line_gpar = list(
list(size = 10, col = "black"),
list(size = 10, col = "black"),
list(size = 10, col = "black")
),
ids = "inner",
label.col = verde_oscuro
) +
geom_node_label(aes(label = paste0("n = ", nodesize)),
ids = "terminal",
size = 3,
nudge_y = 0.01
) +
theme(legend.position = "none")
plot_party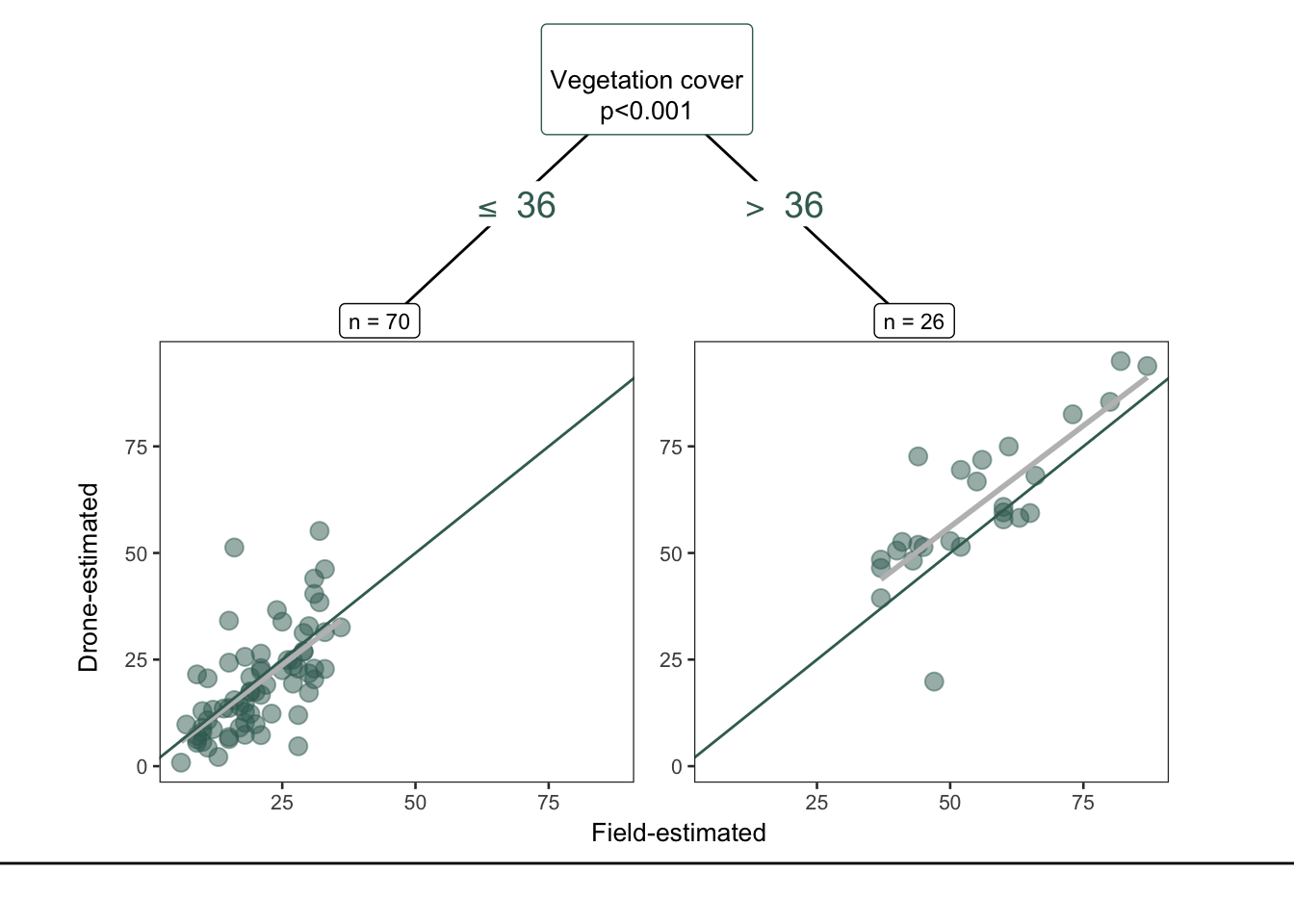
df.rmse_vp <- df %>%
group_by(cover_vp) %>%
summarise(
rmse = round(
Metrics::rmse(plant_coverage_field, plant_coverage_rpas), 4
),
min = min(plant_coverage_field),
max = max(plant_coverage_field),
rmsen.minmax = rmse / (max(plant_coverage_field) - min(plant_coverage_field)) * 100
)
# Compute predictive R2
df.rmse_vp <-
df.rmse_vp |> inner_join(
df |> group_by(cover_vp) |>
group_modify(~ data.frame(
pred.r2 = abs(pred_r_squared(
lm(plant_coverage_rpas ~ plant_coverage_field, data = .x)))))
)| Groups Variance partitioning | RMSE | min | max | norm. RMSE (%) | pred R2 |
|---|---|---|---|---|---|
| cob_high | 11.53 | 37 | 87 | 23.07 | 0.595 |
| cob_low | 9.20 | 6 | 36 | 30.68 | 0.370 |
m <- lm(plant_coverage_rpas ~ plant_coverage_field, data = df)
df <- df %>%
modelr::add_residuals(m) %>%
mutate(resid.abs = abs(resid))
dfres <- df %>%
mutate(abs.Shannon = abs(shannon)) %>%
dplyr::select(
Shannon = abs.Shannon,
Richness = richness, Slope = slope, resid, resid.abs
) %>%
pivot_longer(cols = c("Shannon", "Richness", "Slope")) %>%
mutate(variable = fct_relevel(name, c("Shannon", "Richness", "Slope")))p <- ggpubr::ggscatter(dfres,
x = "value", y = "resid.abs",
color = verde_oscuro,
alpha = 0.5,
xlab = "",
ylab = expression(paste("|", "Residuals", "|")),
add = "reg.line",
add.params = list(color = verde_oscuro, fill = verde_claro),
conf.int = TRUE,
facet.by = "variable"
) +
stat_cor(
label.y.npc = "top", label.x.npc = "left",
aes(label = paste(..rr.label.., ..p.label.., sep = "~`,`~")),
color = verde_oscuro, size = 5
) +
theme(
text = element_text(
colour = verde_oscuro,
size = 12
),
strip.text = element_text(
colour = verde_oscuro,
size = 10
),
axis.title = element_text(size = 12)
)
p.resid <- ggpubr::facet(p,
facet.by = "variable", scales = "free_x",
panel.labs.background = list(fill = "white"),
panel.background = element_blank(),
strip.background = element_blank()
)
p.resid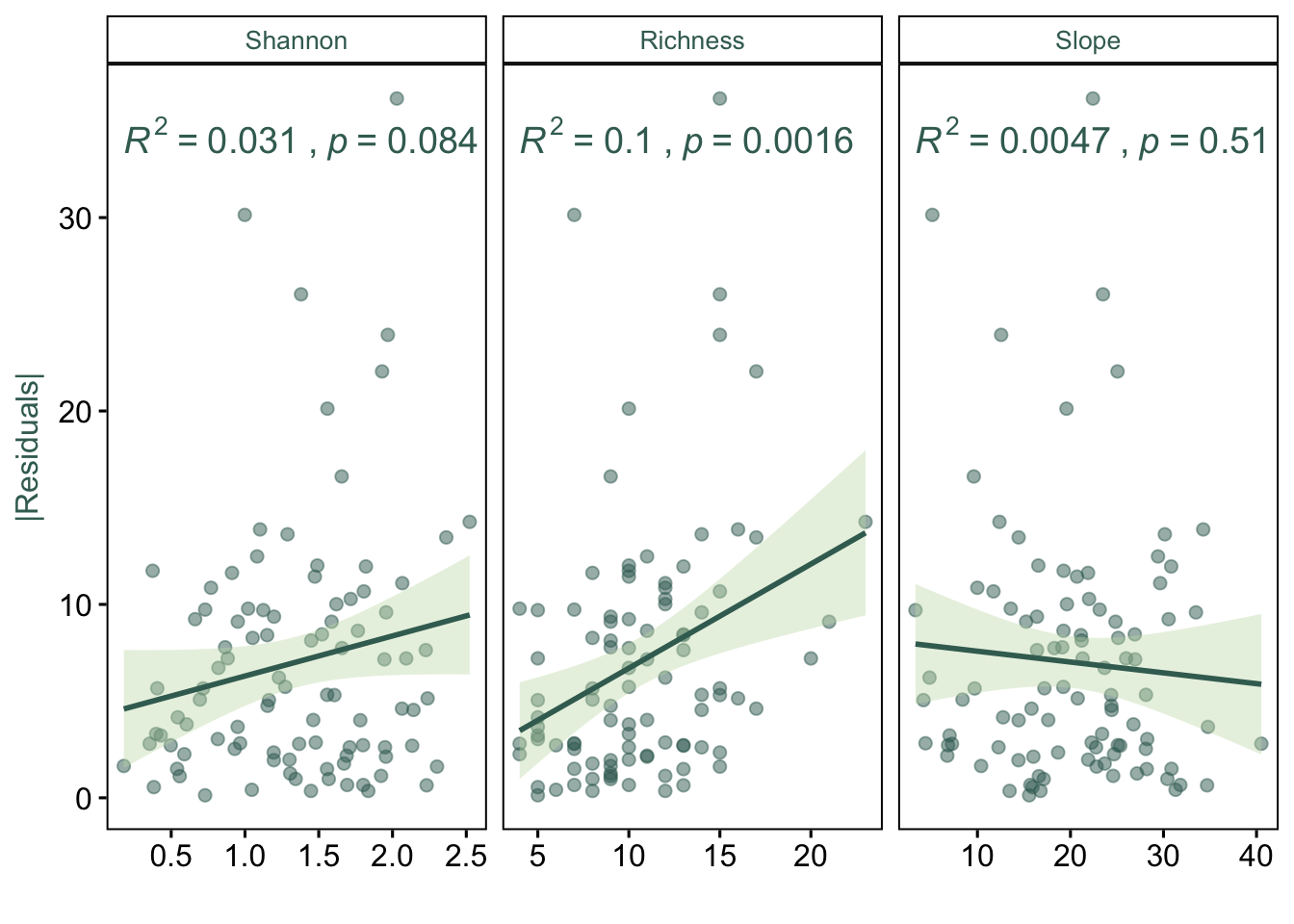
---
title: "Analysis"
editor_options:
chunk_output_type: console
execute:
warning: false
bibliography: grateful-refs.bib
---
```{r}
source("scripts/99-pkgs.R")
```
```{r}
df <- read_excel(
path = here::here("data/rpasfield_alcontar.xlsx"),
sheet = "alcontar"
)
df <- df |>
mutate(cover_type =
case_when(cover_type == "Medium-cover shrubland" ~ "Moderate-cover shrubland",
TRUE ~ cover_type
))
```
## Aims
## General correlation
- Explore the correlation between plant cover estimated by RPAS (RPAS-estimated vegetation cover) and estimated by field measures (Field-estimated vegetation cover)
- Compute the RMSE (Root Mean Squared Error) and the normalized RMSE
```{r}
#| label: rmse-global
df.rmse_global <- df %>%
summarise(
rmse = round(
Metrics::rmse(plant_coverage_field, plant_coverage_rpas), 4
),
min = min(plant_coverage_field),
max = max(plant_coverage_field),
rmsen.minmax = rmse / (max(plant_coverage_field) - min(plant_coverage_field)) * 100
)
```
```{r}
#| label: tab-rmse-global
#| tbl-cap: RMSE and normalized RMSE values for the correlation between the vegetation coverage estimates using RPAS *vs* Field measures
df.rmse_global %>%
kbl(
col.names = c("RMSE", "min", "max", "norm. RMSE (%)"),
digits = 2
) %>%
kable_material()
```
- Compute the LM and the PRESS statistics. More info [here](https://tomhopper.me/2014/05/16/can-we-do-better-than-r-squared/).
```{r}
#| label: tab-lm-global
#| tbl-cap: Estimates of the regression between RPAS and Field measurement
m <- lm(plant_coverage_rpas ~ plant_coverage_field, data = df)
broom::tidy(m) |>
kbl() |>
kable_styling()
```
```{r}
broom::glance(m) |>
t() |>
kbl() |>
kable_styling()
```
The p-value of the regression is `r broom::glance(m)$p.value`.
```{r}
#| label: press-computation
# https://fhernanb.github.io/libro_regresion/diag2.html#estad%C3%ADstica-press-y-r2-de-predicci%C3%B3n
# https://rpubs.com/RatherBit/102428
# https://tomhopper.me/2014/05/16/can-we-do-better-than-r-squared/
PRESS <- function(linear.model) {
# calculate the predictive residuals
pr <- residuals(linear.model) / (1 - lm.influence(linear.model)$hat)
# calculate the PRESS
PRESS <- sum(pr^2)
return(PRESS)
}
pred_r_squared <- function(linear.model) {
#' Use anova() to get the sum of squares for the linear model
lm.anova <- anova(linear.model)
#' Calculate the total sum of squares
tss <- sum(lm.anova$"Sum Sq")
# Calculate the predictive R^2
pred.r.squared <- 1 - PRESS(linear.model) / (tss)
return(pred.r.squared)
}
```
```{r}
PRESS(m)
```
```{r}
pred_r_squared(m)
```
```{r}
# https://stackoverflow.com/questions/17022553/adding-r2-on-graph-with-facets
lm_r2 <- function(df) {
m <- lm(plant_coverage_rpas ~ plant_coverage_field, df)
eq <- substitute(
r2,
list(r2 = format(summary(m)$r.squared, digits = 3))
)
as.character(as.expression(eq))
}
# lm_eqn = function(df, model){
# eq <- substitute(
# italic(y) == a + b %.% italic(x) * "," ~ ~pvalue~p,
# list(
# a = format(coef(model)[1], digits = 3),
# b = format(coef(model)[2], digits = 3),
# p = ifelse(
# summary(m)$coefficients[2,'Pr(>|t|)'] < 0.0001,
# "< 0.0001",
# "= ~format(summary(m)$coefficients[2,'Pr(>|t|)'])"
# )
# ))
# as.character(as.expression(eq))
#
# }
```
```{r}
verde_claro <- "#c6ddb3"
verde_oscuro <- "#3e6c62"
ylab <- "Drone-estimated"
xlab <- "Field-estimated"
# See https://stackoverflow.com/questions/65076492/ggplot-size-of-annotate-vs-size-of-element-text
# Custom theme
theme_rpas <- function() {
theme_bw() %+replace%
theme(
plot.title = element_text(
size = 14,
margin = margin(0, 0, 10, 0)
),
panel.grid = element_blank(),
axis.title = element_text(size = 12),
# For panels
strip.background = element_rect(fill = "white"),
strip.text = element_text(
size = 10,
margin = margin(5, 0, 5, 0)
)
)
}
```
```{r}
formula <- y ~ x
general_plot <- df %>%
ggplot(aes(x = plant_coverage_field, y = plant_coverage_rpas)) +
geom_point(size = 3, alpha = .5, colour = verde_oscuro) +
geom_abline(slope = 1, colour = verde_oscuro) +
labs(
x = xlab, y = ylab,
title = "Vegetation cover (%)"
) +
xlim(0, 100) +
ylim(0, 100) +
stat_poly_eq(
formula = formula,
aes(label = paste(
after_stat(eq.label), "*\"; \"*",
after_stat(adj.rr.label), "*\"; \"*",
after_stat(p.value.label), "*\"\"",
sep = ""
)),
rr.digits = 3,
colour = "black",
size = 8 / .pt
) +
annotate("text",
x = 0, y = 85,
label = paste0("RMSE = ", round(df.rmse_global$rmse, 2)),
colour = "black",
size = 12 / .pt,
hjust = 0
) +
annotate("text",
x = 0, y = 75,
label = paste0(
"italic(R)[predic.]^2~'='~",
round(pred_r_squared(m), 3)
),
colour = "black",
parse = TRUE,
size = 12 / .pt,
hjust = 0
) +
theme_rpas() +
geom_smooth(method = "lm", col = "gray", se = FALSE, size = 1)
general_plot
```
## Comparison by treatments
- Is the estimation different between treatments?
```{r}
#| label: rmse-treatment
df.rmse_treatment <- df %>%
group_by(treatment) %>%
summarise(
rmse = round(
Metrics::rmse(plant_coverage_field, plant_coverage_rpas), 4
),
min = min(plant_coverage_field),
max = max(plant_coverage_field),
rmsen.minmax = rmse / (max(plant_coverage_field) - min(plant_coverage_field)) * 100
)
# Compute predictive R2
df.rmse_treatment <-
df.rmse_treatment |> inner_join(
df |> group_by(treatment) |>
group_modify(~ data.frame(
pred.r2 = pred_r_squared(
lm(plant_coverage_rpas ~ plant_coverage_field, data = .x))))
)
```
```{r}
#| label: tab-rmse-treatment
#| tbl-cap: RMSE and normalized RMSE values for the correlation between the vegetation coverage estimates using RPAS *vs* Field measures. Values by treatment
df.rmse_treatment %>%
kbl(
col.names = c("Treatment", "RMSE", "min", "max", "norm. RMSE (%)", "pred R2"),
digits = c(0, 2, 0, 0, 2,3)
) %>%
kable_material()
```
```{r}
eqns <- by(df, df$treatment, lm_r2)
df.label <- data.frame(eq = unclass(eqns), treatment = names(eqns))
df.label$lab <- paste(df.label$treatment, "R^2 =", df.label$eq, sep = " ")
r2_labeller <- function(variable, value) {
return(df.label$lab)
}
```
```{r}
treatment_plot <- df %>%
ggplot(aes(x = plant_coverage_field, y = plant_coverage_rpas)) +
geom_point(size = 3, alpha = .5, colour = verde_oscuro) +
geom_abline(slope = 1, colour = verde_oscuro) +
facet_wrap(~treatment, labeller = label_value) +
labs(x = xlab, y = ylab, title = "Vegetation cover (%)") +
xlim(0, 100) +
ylim(0, 100) +
theme_rpas() +
theme(legend.position = "none") +
geom_richtext(
data = df.rmse_treatment, size = 8 / .pt,
aes(
x = 0, y = 80,
label = paste0(
"RMSE<sub>norm.</sub> = ",
round(rmsen.minmax, 2), " %"
),
hjust = 0
),
fill = NA, label.color = NA
) +
stat_poly_eq(
formula = formula,
aes(label = paste(
after_stat(adj.rr.label), "*\"; \"*",
after_stat(p.value.label), "*\"\"",
sep = ""
)),
rr.digits = 3,
colour = "black",
size = 8 / .pt
) +
geom_richtext(
data = df.rmse_treatment, size = 8 / .pt,
aes(
x = 0, y = 70,
label = paste0(
"*R*<sup>2</sup><sub>predic.</sub> = ",
round(pred.r2, 3)
),
hjust = 0
),
fill = NA, label.color = NA
) +
geom_smooth(method = "lm", col = "gray", se = FALSE, size = 1)
treatment_plot
```
## Comparison by cover types
```{r}
#| label: rmse-groups
df.rmse_groups <- df %>%
group_by(cover_type) %>%
summarise(
rmse = round(
Metrics::rmse(plant_coverage_field, plant_coverage_rpas), 4
),
min = min(plant_coverage_field),
max = max(plant_coverage_field),
rmsen.minmax = rmse / (max(plant_coverage_field) - min(plant_coverage_field)) * 100
)
# Compute predictive R2
df.rmse_groups <-
df.rmse_groups |> inner_join(
df |> group_by(cover_type) |>
group_modify(~ data.frame(
pred.r2 = abs(pred_r_squared(
lm(plant_coverage_rpas ~ plant_coverage_field, data = .x)))))
)
```
```{r}
#| label: tab-rmse-groups
#| tbl-cap: RMSE and normalized RMSE values for the correlation between the vegetation coverage estimates using RPAS *vs* Field measures. Values by cover types
df.rmse_groups %>%
kbl(
col.names = c("Cover type", "RMSE", "min", "max", "norm. RMSE (%)", "pred R2"),
digits = c(0, 2, 0, 0, 2,3)
) %>%
kable_material()
```
```{r}
eqns <- by(df, df$cover_type, lm_r2)
df.label <- data.frame(eq = unclass(eqns), treatment = names(eqns))
df.label$lab <- paste(df.label$cover_type, "R^2 =", df.label$eq, sep = " ")
r2_labeller <- function(variable, value) {
return(df.label$lab)
}
```
```{r}
custom_order <- c(
"Low-cover shrubland", "Moderate-cover shrubland",
"High-cover shrubland", "High-cover alfa grass steppe"
)
covertype_plot <- df %>%
ggplot(aes(x = plant_coverage_field, y = plant_coverage_rpas)) +
geom_point(size = 3, alpha = .5, colour = verde_oscuro) +
geom_abline(slope = 1, colour = verde_oscuro) +
facet_wrap(~ factor(cover_type, custom_order),
labeller = label_value
) +
labs(x = xlab, y = ylab, title = "Vegetation cover (%)") +
xlim(0, 100) +
ylim(0, 100) +
theme_rpas() +
theme(legend.position = "none") +
geom_richtext(
data = df.rmse_groups, size = 8 / .pt,
aes(
x = 0, y = 80,
label = paste0(
"RMSE<sub>norm.</sub> = ",
round(rmsen.minmax, 2), " %"
)
),
hjust = 0,
fill = NA, label.color = NA
) +
stat_poly_eq(
formula = formula,
aes(label = paste(
after_stat(adj.rr.label), "*\"; \"*",
after_stat(p.value.label), "*\"\"",
sep = ""
)),
rr.digits = 3,
colour = "black",
size = 8 / .pt
) +
geom_richtext(
data = df.rmse_groups, size = 8 / .pt,
aes(
x = 0, y = 70,
label = paste0(
"*R*<sup>2</sup><sub>predic.</sub> = ",
round(pred.r2, 3)
),
hjust = 0
),
fill = NA, label.color = NA
) +
geom_smooth(method = "lm", col = "gray", se = FALSE, size = 1)
covertype_plot
```
```{r}
combined_plot <- covertype_plot / treatment_plot +
plot_layout(heights = c(2, 1)) +
plot_annotation(tag_levels = "a") &
theme(plot.tag = element_text(face = "bold"))
```
## Variance partitioning
Is the relationship between the vegetation coverage estimated by RPAS and by field measurement uniform across all coverage values? or is the correlation between those two approach homogeneous across all coverage values?
```{r}
ct <- partykit::ctree(plant_coverage_rpas ~ plant_coverage_field, data = df)
ct
sctest(ct)
```
There are 5 terminal nodes.
```{r}
ggparty(ct) +
geom_edge() +
geom_edge_label(colour = "grey", size = 4) +
geom_node_plot(
gglist = list(
geom_point(aes(x = plant_coverage_field, y = plant_coverage_rpas)),
geom_smooth(aes(x = plant_coverage_field, y = plant_coverage_rpas), method = lm, se = FALSE),
geom_abline(slope = 1, colour = "gray"),
theme_bw(base_size = 10),
xlab(xlab),
ylab(ylab)
),
scales = "fixed",
shared_axis_labels = TRUE,
shared_legend = TRUE,
legend_separator = TRUE,
id = "terminal"
) +
geom_node_label(aes(col = splitvar),
line_list = list(
aes(label = paste("Node", id)),
aes(label = splitvar),
aes(label = scales::pvalue(p.value,
accuracy = 0.001,
decimal.mark = ".",
add_p = TRUE
))
),
line_gpar = list(
list(size = 8, col = "black", fontface = "bold"),
list(size = 8),
list(size = 8)
),
ids = "inner"
) +
geom_node_label(aes(label = paste0("Node ", id, ", (n= ", nodesize, ")")),
fontface = "bold",
ids = "terminal",
size = 2,
nudge_y = 0.01
) +
theme(legend.position = "none")
```
What about the overfitting?
```{r}
ggplot(df, aes(x = plant_coverage_field, y = plant_coverage_rpas)) +
geom_point() +
geom_abline(yintercept = 1) +
geom_smooth(
data = (df %>% filter(plant_coverage_field <= 13)),
aes(x = plant_coverage_field, y = plant_coverage_rpas),
method = "lm"
) +
geom_smooth(
data = (df %>% filter(plant_coverage_field > 13 & plant_coverage_field <= 23)),
aes(x = plant_coverage_field, y = plant_coverage_rpas),
method = "lm"
) +
geom_smooth(
data = (df %>% filter(plant_coverage_field > 23 & plant_coverage_field <= 36)),
aes(x = plant_coverage_field, y = plant_coverage_rpas),
method = "lm"
) +
geom_smooth(
data = (df %>% filter(plant_coverage_field > 36 & plant_coverage_field <= 50)),
aes(x = plant_coverage_field, y = plant_coverage_rpas),
method = "lm"
) +
geom_smooth(
data = (df %>% filter(plant_coverage_field > 50)),
aes(x = plant_coverage_field, y = plant_coverage_rpas),
method = "lm"
)
```
### Find the optimous complexity parameter
```{r}
set.seed(123)
ctrpart <- rpart(plant_coverage_rpas ~ plant_coverage_field, data = df)
ctrpart
printcp(ctrpart)
plotcp(ctrpart)
```
```{r}
set.seed(123)
ctrpart2 <- rpart(plant_coverage_rpas ~ plant_coverage_field,
data = df, control =
rpart.control(minsplit = 2, cp = .075)
)
ctrpart2
printcp(ctrpart2)
plotcp(ctrpart2)
```
```{r}
set.seed(123)
ct_ok <- partykit::ctree(plant_coverage_rpas ~ plant_coverage_field,
data = df,
control =
ctree_control(minsplit = 1, alpha = 0.05, maxdepth = 1)
)
plot_party <- ggparty(ct_ok) +
geom_edge() +
geom_edge_label(colour = verde_oscuro, size = 5) +
geom_node_plot(
gglist = list(
geom_point(
aes(
x = plant_coverage_field,
y = plant_coverage_rpas
),
size = 3, alpha = .5, colour = verde_oscuro
),
geom_smooth(aes(x = plant_coverage_field, y = plant_coverage_rpas),
method = lm, se = FALSE, colour = "gray"
),
geom_abline(slope = 1, colour = verde_oscuro),
theme_bw(base_size = 10),
xlab(xlab),
ylab(ylab),
theme(
panel.grid = element_blank()
)
),
scales = "fixed",
shared_axis_labels = TRUE,
shared_legend = TRUE,
legend_separator = TRUE,
id = "terminal"
) +
geom_node_label(aes(col = splitvar),
line_list = list(
aes(id),
aes(label = "Vegetation cover"),
aes(label = scales::pvalue(p.value,
accuracy = 0.001,
decimal.mark = ".",
add_p = TRUE
))
),
line_gpar = list(
list(size = 10, col = "black"),
list(size = 10, col = "black"),
list(size = 10, col = "black")
),
ids = "inner",
label.col = verde_oscuro
) +
geom_node_label(aes(label = paste0("n = ", nodesize)),
ids = "terminal",
size = 3,
nudge_y = 0.01
) +
theme(legend.position = "none")
plot_party
```
```{r}
df <- df %>%
mutate(cover_vp = case_when(
plant_coverage_field <= 36 ~ as.character("cob_low"),
TRUE ~ "cob_high"
))
```
```{r}
#| label: rmse-vp
df.rmse_vp <- df %>%
group_by(cover_vp) %>%
summarise(
rmse = round(
Metrics::rmse(plant_coverage_field, plant_coverage_rpas), 4
),
min = min(plant_coverage_field),
max = max(plant_coverage_field),
rmsen.minmax = rmse / (max(plant_coverage_field) - min(plant_coverage_field)) * 100
)
# Compute predictive R2
df.rmse_vp <-
df.rmse_vp |> inner_join(
df |> group_by(cover_vp) |>
group_modify(~ data.frame(
pred.r2 = abs(pred_r_squared(
lm(plant_coverage_rpas ~ plant_coverage_field, data = .x)))))
)
```
```{r}
#| label: tab-rmse-vp
#| tbl-cap: RMSE and normalized RMSE values for the correlation between the vegetation coverage estimates using RPAS *vs* Field measures. Values by variance partitioning-groups.
df.rmse_vp %>%
kbl(
col.names = c("Groups Variance partitioning", "RMSE", "min", "max", "norm. RMSE (%)", "pred R2"),
digits = c(0, 2, 0, 0, 2,3)
) %>%
kable_material()
```
# Is the the estimation influenced by other variables?
```{r}
m <- lm(plant_coverage_rpas ~ plant_coverage_field, data = df)
df <- df %>%
modelr::add_residuals(m) %>%
mutate(resid.abs = abs(resid))
dfres <- df %>%
mutate(abs.Shannon = abs(shannon)) %>%
dplyr::select(
Shannon = abs.Shannon,
Richness = richness, Slope = slope, resid, resid.abs
) %>%
pivot_longer(cols = c("Shannon", "Richness", "Slope")) %>%
mutate(variable = fct_relevel(name, c("Shannon", "Richness", "Slope")))
```
```{r}
p <- ggpubr::ggscatter(dfres,
x = "value", y = "resid.abs",
color = verde_oscuro,
alpha = 0.5,
xlab = "",
ylab = expression(paste("|", "Residuals", "|")),
add = "reg.line",
add.params = list(color = verde_oscuro, fill = verde_claro),
conf.int = TRUE,
facet.by = "variable"
) +
stat_cor(
label.y.npc = "top", label.x.npc = "left",
aes(label = paste(..rr.label.., ..p.label.., sep = "~`,`~")),
color = verde_oscuro, size = 5
) +
theme(
text = element_text(
colour = verde_oscuro,
size = 12
),
strip.text = element_text(
colour = verde_oscuro,
size = 10
),
axis.title = element_text(size = 12)
)
p.resid <- ggpubr::facet(p,
facet.by = "variable", scales = "free_x",
panel.labs.background = list(fill = "white"),
panel.background = element_blank(),
strip.background = element_blank()
)
p.resid
```
```{r}
#| echo: false
#| label: generate-jpg
ggsave(general_plot,
filename = here::here("output/correla_general.jpg"),
device = "jpg", height = 10, width = 10, unit = "cm", dpi = "print"
)
ggsave(treatment_plot,
filename = here::here("output/correla_treatment.jpg"),
device = "jpg", height = 10, width = 15, unit = "cm", dpi = "print"
)
ggsave(covertype_plot,
filename = here::here("output/correla_covertype.jpg"),
device = "jpg", height = 15, width = 15, unit = "cm", dpi = "print"
)
ggsave(combined_plot,
filename = here::here("output/correla_combined.jpg"),
device = "jpg", height = 20, width = 14, unit = "cm", dpi = "print"
)
ggsave(p.resid,
filename = here::here("output/residuals.jpg"),
device = "jpg", height = 8, width = 20, unit = "cm", dpi = "print"
)
ggsave(plot_party,
filename = here::here("output/correla_partitioning.jpg"),
device = "jpg", height = 12, width = 12, unit = "cm", dpi = "print", bg = "white"
)
ggsave(plot_party,
filename = here::here("output/correla_partitioning.svg"),
device = "svg", height = 12, width = 12, unit = "cm", dpi = "print", bg = "white"
)
# PNG devices
ggsave(general_plot,
filename = here::here("output/correla_general_transp.png"),
device = "png", height = 10, width = 10, unit = "cm", dpi = "print",
bg = "transparent"
)
ggsave(treatment_plot,
filename = here::here("output/correla_treatment_transp.png"),
device = "png", height = 10, width = 15, unit = "cm", dpi = "print",
bg = "transparent"
)
ggsave(covertype_plot,
filename = here::here("output/correla_covertype_transp.png"),
device = "png", height = 15, width = 15, unit = "cm", dpi = "print",
bg = "transparent"
)
ggsave(combined_plot,
filename = here::here("output/correla_combined.png"),
device = "png", height = 20, width = 14, unit = "cm", dpi = "print",
bg = "transparent"
)
ggsave(p.resid,
filename = here::here("output/residuals.png"),
device = "png", height = 8, width = 20, unit = "cm", dpi = "print",
bg = "transparent"
)
ggsave(plot_party,
filename = here::here("output/correla_partitioning.jpg"),
device = "png", height = 12, width = 12, unit = "cm", dpi = "print",
bg = "transparent"
)
## SVG
ggsave(general_plot,
filename = here::here("output/correla_general.svg"),
device = "svg", height = 10, width = 10, unit = "cm", dpi = "print"
)
ggsave(treatment_plot,
filename = here::here("output/correla_treatment.svg"),
device = "svg", height = 10, width = 15, unit = "cm", dpi = "print"
)
ggsave(covertype_plot,
filename = here::here("output/correla_covertype.svg"),
device = "svg", height = 15, width = 15, unit = "cm", dpi = "print"
)
ggsave(combined_plot,
filename = here::here("output/correla_combined.svg"),
device = "svg", height = 20, width = 14, unit = "cm", dpi = "print"
)
ggsave(p.resid,
filename = here::here("output/residuals.svg"),
device = "svg", height = 8, width = 20, unit = "cm", dpi = "print"
)
ggsave(plot_party,
filename = here::here("output/correla_partitioning.svg"),
device = "svg", height = 12, width = 12, unit = "cm", dpi = "print", bg = "white"
)
```